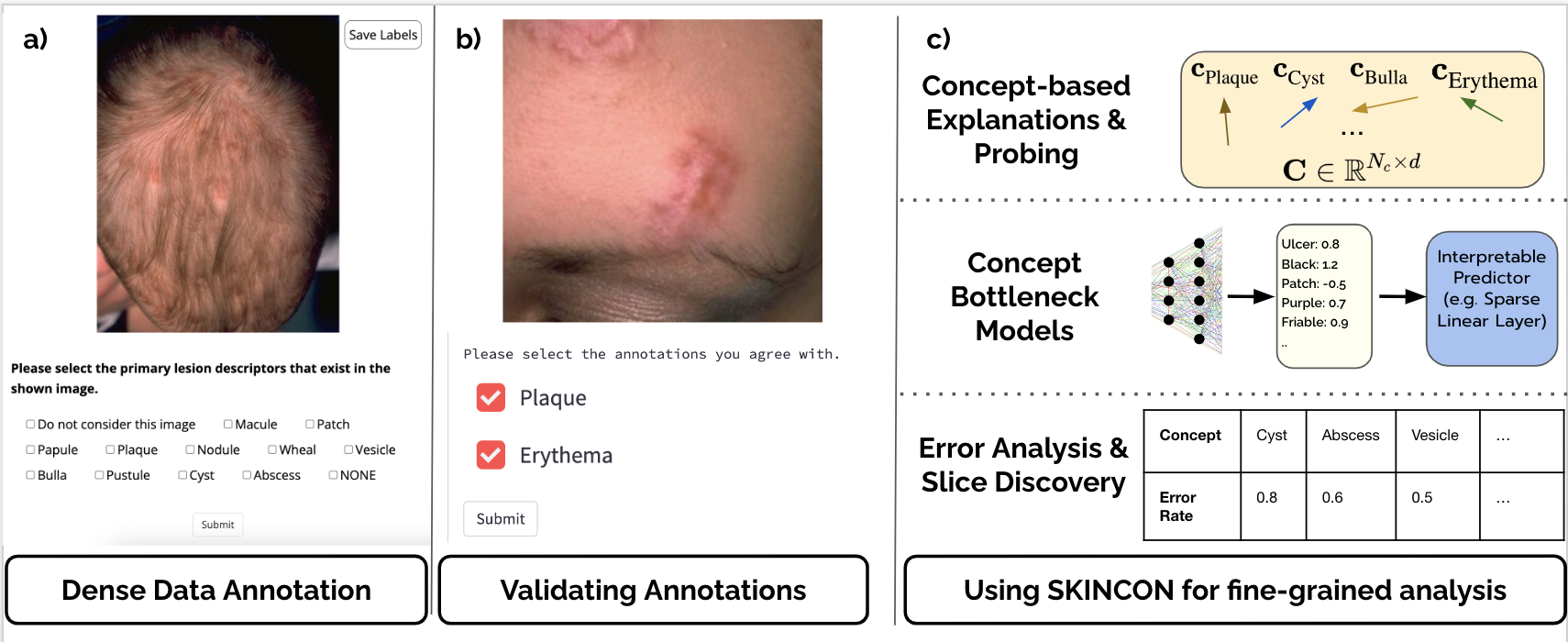
For the deployment of artificial intelligence (AI) in high risk settings, such as healthcare, methods that provide interpretability/explainability or allow fine-grained error analysis are critical. Many recent methods for interpretability/explainability and fine-grained error analysis use concepts, which are meta-labels which are semantically meaningful to humans. However, there are only a few datasets that include concept-level meta-labels and most of these meta-labels are relevant for natural images that do not require domain expertise. Previous densely annotated datasets in medicine focused on meta-labels that are relevant to a single disease such as osteoarthritis or melanoma. In dermatology, skin disease is described using an established clinical lexicon that allow clinicians to describe physical exam findings to one another. To provide the first medical dataset densely annotated by domain experts to provide annotations useful across multiple disease processes, we developed SKINCON: a skin disease dataset densely annotated by dermatologists. SKINCON includes 3230 images from the Fitzpatrick 17k skin disease dataset densely annotated with 48 clinical concepts, 22 of which have at least 50 images representing the concept. The concepts used were chosen by two dermatologists considering the clinical descriptor terms used to describe skin lesions. Examples include "plaque", "scale", and "erosion". These same concepts were also used to label 656 skin disease images from the Diverse Dermatology Images dataset, providing an additional external dataset with diverse skin tone representations. We review the potential applications for the SKINCON dataset, such as probing models, concept-based explanations, concept bottlenecks, error analysis, and slice discovery. Furthermore, we use SKINCON to demonstrate two of these use cases: probing an existing dermatology AI model for concepts with concept activation vectors and developing interpretable models with post-hoc concept bottleneck models.